DRPScore & DDPScore
Evaluation of RNA/DNA-protein complex structures using the deep learning method

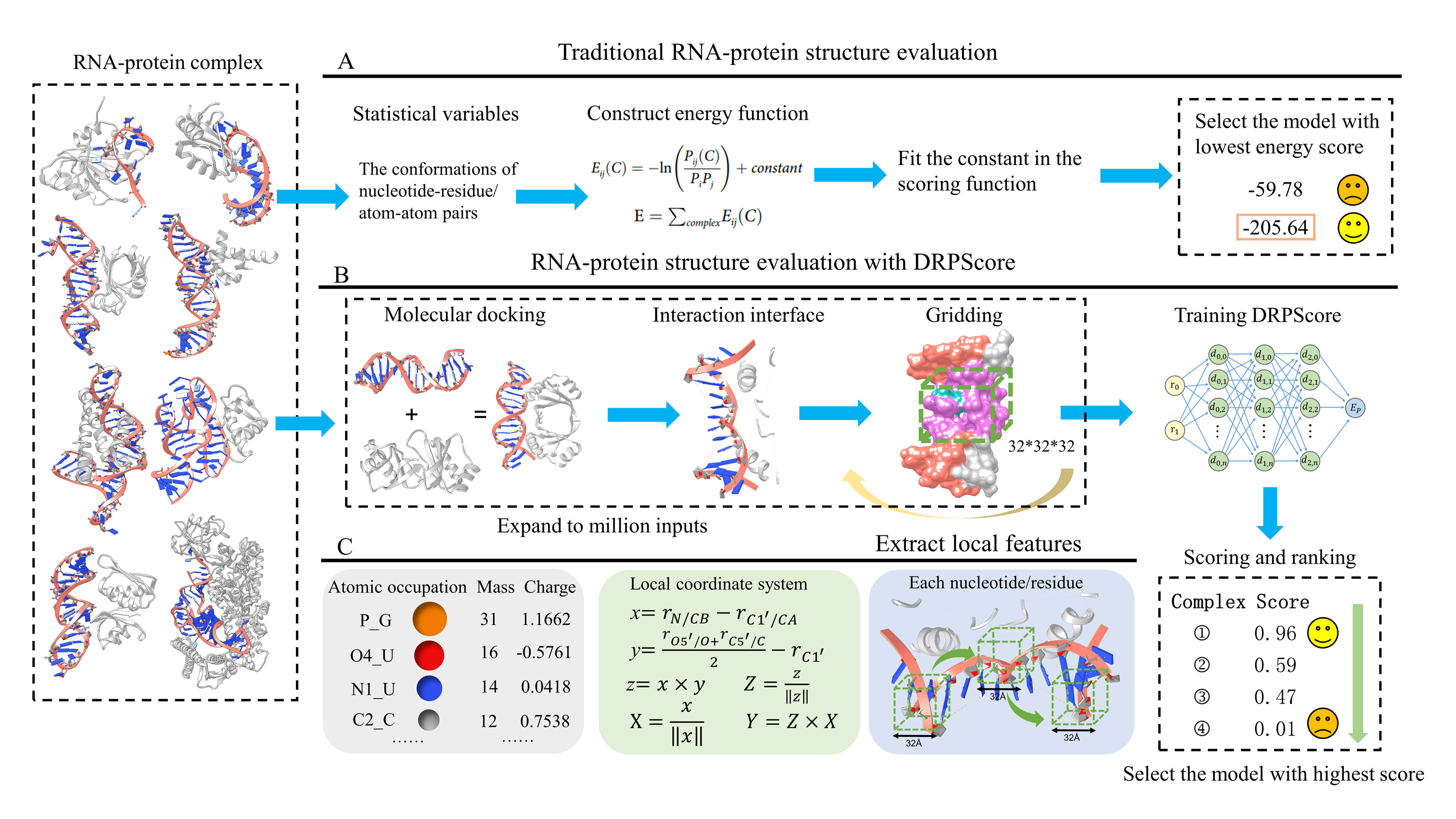
About DRPScore
RNA-protein complexes are fundamental to numerous cellular processes, including translation and gene regulation. The high-resolution structural determination of RNA-protein complexes is essential to understanding their functions. Therefore, computational methods capable of identifying native-like RNA-protein structures are needed. To address this challenge, we have developed DRPScore, a deep-learning-based approach for identifying native-like RNA-protein structures.
DRPScore has been tested on representative sets of RNA-protein complexes that exhibit various degrees of binding-induced conformation changes. These range from fully rigid docking (bound-bound) to fully flexible docking (unbound-unbound). Out of the top 20 predictions, DRPScore selects native-like structures with a success rate of 91.67% for bound RNA-protein complexes and 56.14% for unbound complexes. DRPScore consistently outperforms existing methods, achieving a 10.53-15.79% improvement, even in the most challenging unbound cases. Furthermore, DRPScore significantly enhances the accuracy of native interface interaction predictions. DRPScore is expected to be widely useful in modeling and designing RNA-protein complexes.
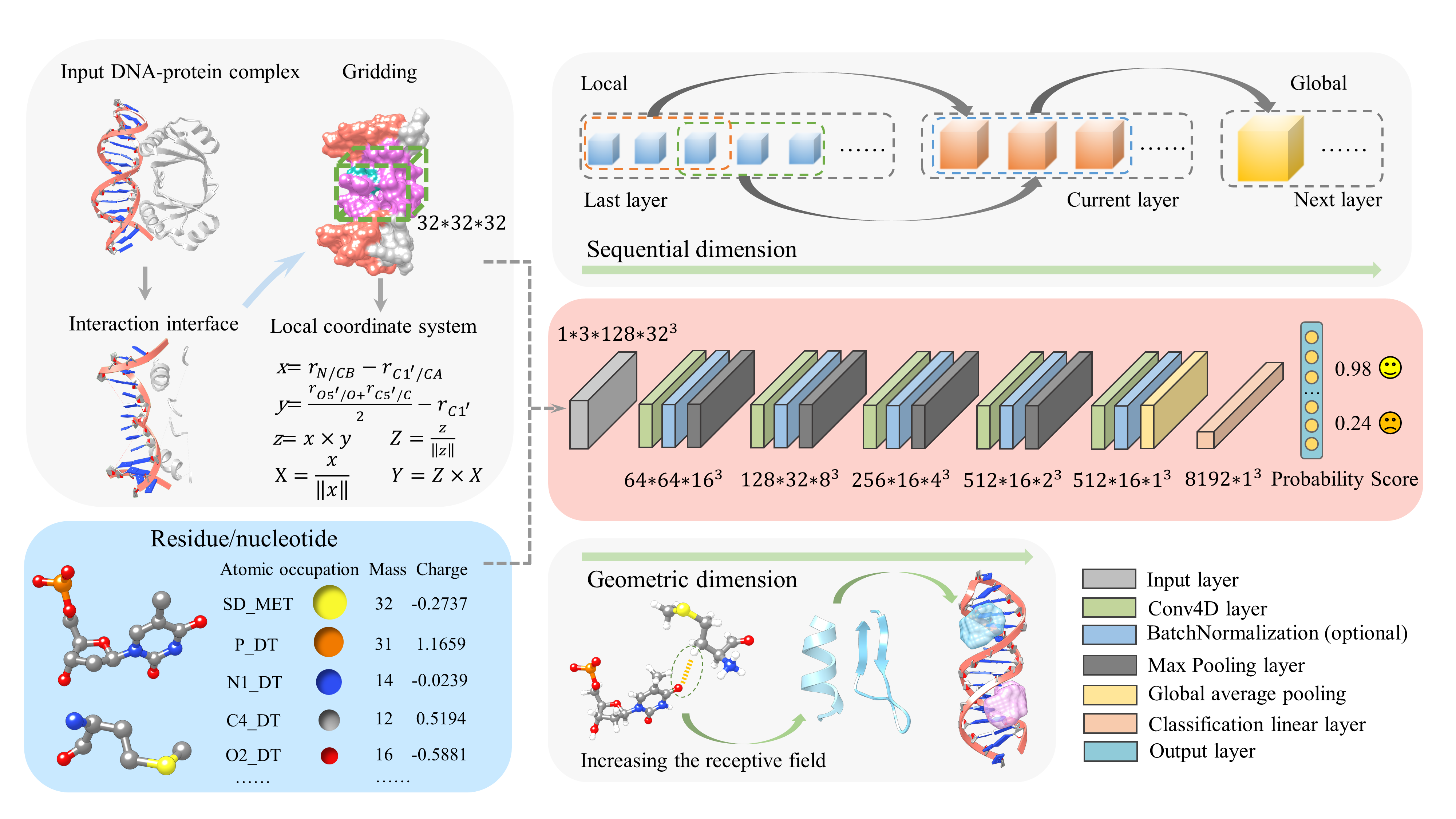
About DDPScore
Biological processes such as transcription, repair, and regulation require interactions between DNA and proteins. To unravel their functions, it is imperative to determine the high-resolution structures of DNA-protein complexes. However, experimental methods for this purpose are costly and technically demanding. Consequently, there is an urgent need for computational techniques to identify the structures of DNA-protein complexes. Despite technological advancements, accurately identifying DNA-protein complexes through computational methods still poses a challenge.
Our team has developed a cutting-edge deep-learning approach called DDPScore that assesses DNA-protein complex structures. DDPScore utilizes a 4D convolutional neural network to overcome limited training data. This approach effectively captures local and global features while comprehensively considering the conformational changes arising from the flexibility during the DNA-protein docking process. DDPScore consistently outperformed the available methods in comprehensive DNA-protein complex docking evaluations, even for the flexible docking challenges. DDPScore has a wide range of applications in predicting and designing structures of DNA-protein complexes.
Resources
Download DDPScore V1.1 Standalone Package
Download the Available RNA-protein Complex Data
Download the Available DNA-protein Complex Data
Related Links
I. 3D Structure Resources
II. RNA Structure Prediction
III. Molecular Docking
IV. Molecular Dynamics
V. Molecular Visualization/Analysis
Visitor Information
| Region | Visit Count | IP |
|---|
Contact
If you have any questions, feel free to contact us at: yjzhaowh@mail.ccnu.edu.cn
Institute of Biophysics and Department of Physics, Central China Normal University, Wuhan 430079, China.
References
- Chengwei Zeng#, Yiren Jian#, Soroush Vosoughi, Chen Zeng, and Yunjie Zhao*, Evaluating native-like structures of RNA-protein complexes through the deep learning method. Nature Communications, 2023. 14(1):1060.
- Chengwei Zeng#, Yiren Jian#, Chen Zhuo, Anbang Li, Chen Zeng and Yunjie Zhao*, Evaluation of DNA-protein complex structures using the deep learning method, Physical Chemistry Chemical Physics, 2024. 26(1):130-143.