The output example of an input example (2ZNI_RNA_map.pdb)
1. RNA binding site prediction results
In the network, a node represents single nucleotide, and the non-covalent interactions between nodes separated by less than 8 Å forming the edges. The radius of a node is proportional to the closeness value for the corresponding nucleotide. The red nodes are the predicted binding sites whose names, closeness, and degree values are listed in the table on the right. Users can get the nucleotide name, nucleotide number, closeness, and degree values if put the mouse at the node. Moreover, users can zoom the network by scrolling the mouse and drag the network for particularly detailed network information.
2. The statistical analysis of the closeness and degree calculation results.
The closeness histogram: The X-axis shows the closeness and the Y-axis shows the number of nucleotides.
The degree histogram: The X-axis shows the degree and the Y-axis shows the number of nucleotides.
2.1 Closeness_histogram
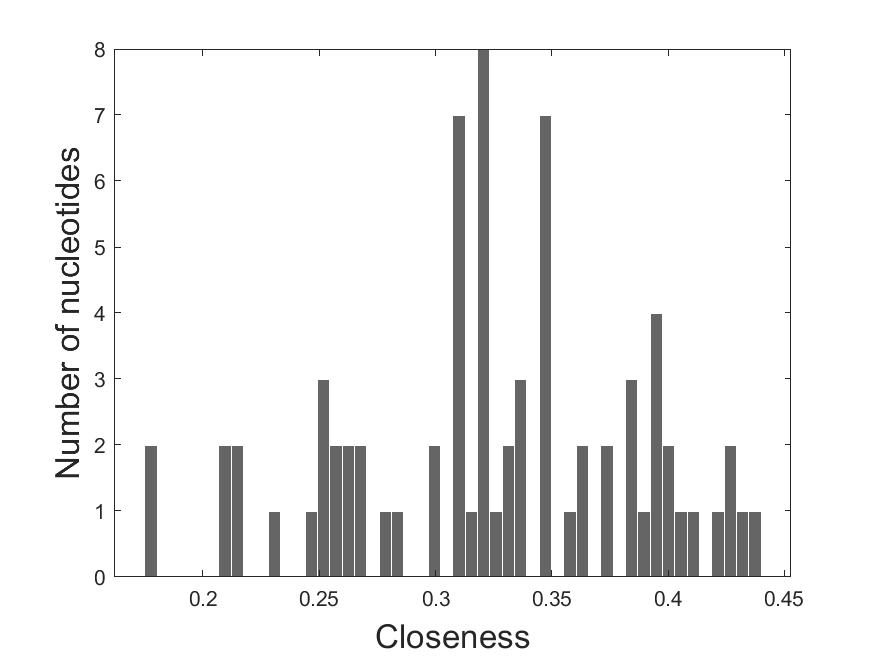
2.2 Degree_histogram
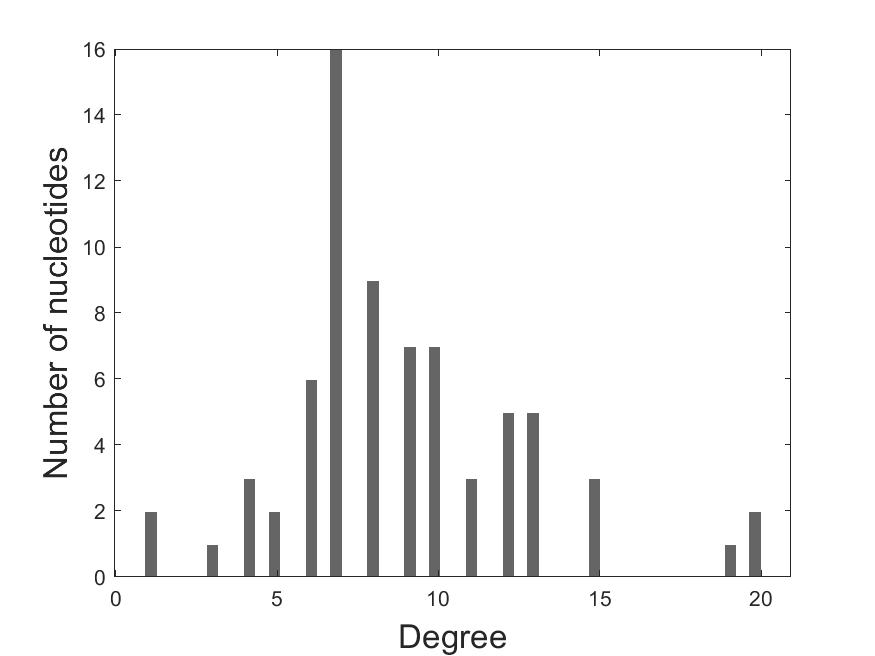
3. Download Results
The results consisted of three modules: nodes, edges, and sites.
1. The "nodes" module contains the nucleotide names, closeness and degree values for network nodes.
2. The "edges" module contains the information of connected nodes.
3. The "sites" module contains nucleotide name, closeness and degree values for predicted binding sites.
1. The "nodes" module contains the nucleotide names, closeness and degree values for network nodes.
2. The "edges" module contains the information of connected nodes.
3. The "sites" module contains nucleotide name, closeness and degree values for predicted binding sites.