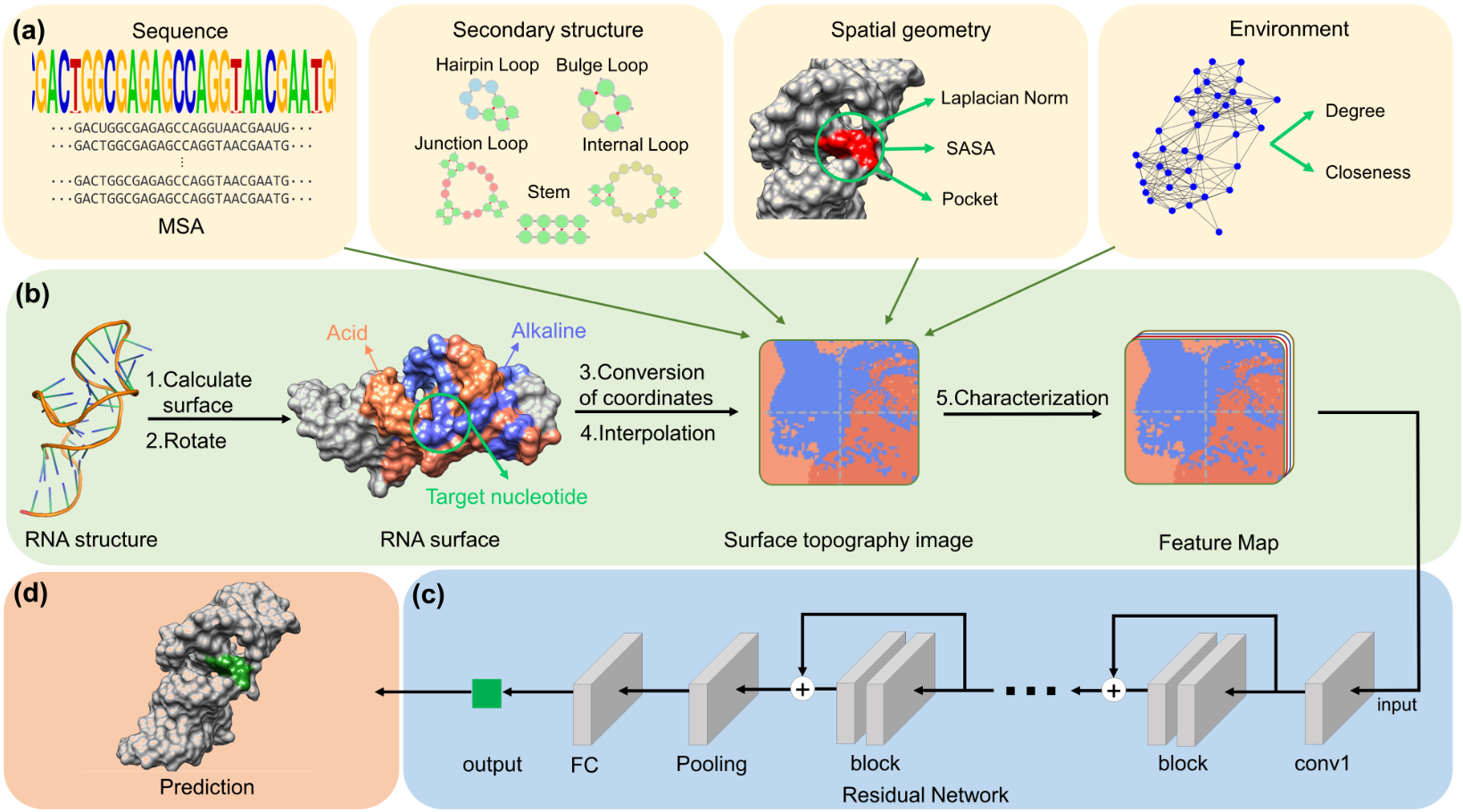
RNA-small molecule interactions play a crucial role in drug discovery and inhibitor design. Identifying RNA-small molecule binding nucleotides is essential and requires methods that exhibit high predictive ability to facilitate drug discovery and inhibitor design. Existing methods can predict the binding nucleotides of simple RNA structures, but it is hard to predict binding nucleotides in complex RNA structures with junctions. To address this limitation, we developed a new deep learning model based on spatial correlation, ZHmolReSTasite, which can accurately predict binding nucleotides of small and large RNA with junctions. We utilize RNA surface topography to consider the spatial correlation, characterizing nucleotides from sequence and tertiary structure to learn a high-level representation. Our method outperforms existing methods for benchmark test sets composed of simple RNA structures, achieving precision values of 72.9% on TE18 and 76.7% on RB9 test sets. For a challenging test set composed of RNA structures with junctions, our method outperforms the second-best method by 11.6% in precision. Moreover, ZHmolReSTasite demonstrates robustness regarding predicted RNA structures. In summary, ZHmolReSTasite successfully incorporates spatial correlation, outperforms previous methods on small and large RNA structures using RNA surface topography, and can provide valuable insights into RNA-small molecule prediction and accelerate RNA inhibitor design.
